Epitope mapping of mono- and polyclonal antibodies by screening phage-displayed random peptide libraries
DOI:
https://doi.org/10.17344/acsi.2016.2458Keywords:
peptide, epitope mapping, mimotope, phage display, antibodyAbstract
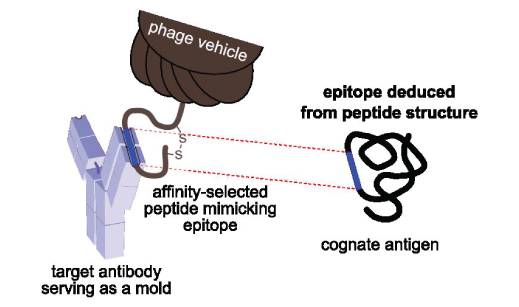
Detailed knowledge of antigenic determinants is crucial when characterizing therapeutic and diagnostic antibodies, assessing vaccine effectiveness and developing epitope-based vaccines. Most epitope mapping approaches are labor intensive and costly. In this study, we evaluated panning of phage-displayed random peptide libraries against antibodies as a tool for cognate epitope identification. We used six antibodies directed to three model protein antigens as targets to show that the approach is applicable to both mono- and polyclonal antibodies. The technique is well-suited especially for identification of linear epitopes. Mapping of conformational epitopes is more challenging, tends to be more subjective and requires use of computational tools. Nevertheless, when combined with functional data such as structure-activity relationship of antigen muteins, one can make reliable conformational epitope predictions based on phage display experiment data. As the described approach is fast and relatively inexpensive, we suggest it is employed early in antibody characterization and later validated by complementary methods.
Downloads
Additional Files
Published
Issue
Section
License
Except where otherwise noted, articles in this journal are published under the Creative Commons Attribution 4.0 International License
